Technology
TargetScan
At the heart of the TScan’s discovery process is our proprietary genome-wide, high-throughput target identification screen, designed to rapidly identify the natural targets of T cell receptors.
The TargetScan discovery platform enables the identification of the natural target of a T cell receptor, or TCR, using an unbiased, genome-wide, high-throughput screen. We have developed this technology to be extremely versatile and applicable across multiple therapeutic areas, including cancer, autoimmune disorders, and infectious diseases. It can be applied to virtually any TCR that plays a role in the cause or prevention of disease. TargetScan is also designed to identify potential off-targets of a TCR and eliminate those TCR candidates that cross-react with proteins expressed at high levels in critical organs. We believe this will allow us to reduce the risk and enhance the potential safety profile of our TCR-T therapy candidates early in development before we initiate clinical trials.
Overview of Our Proprietary TargetScan Technology
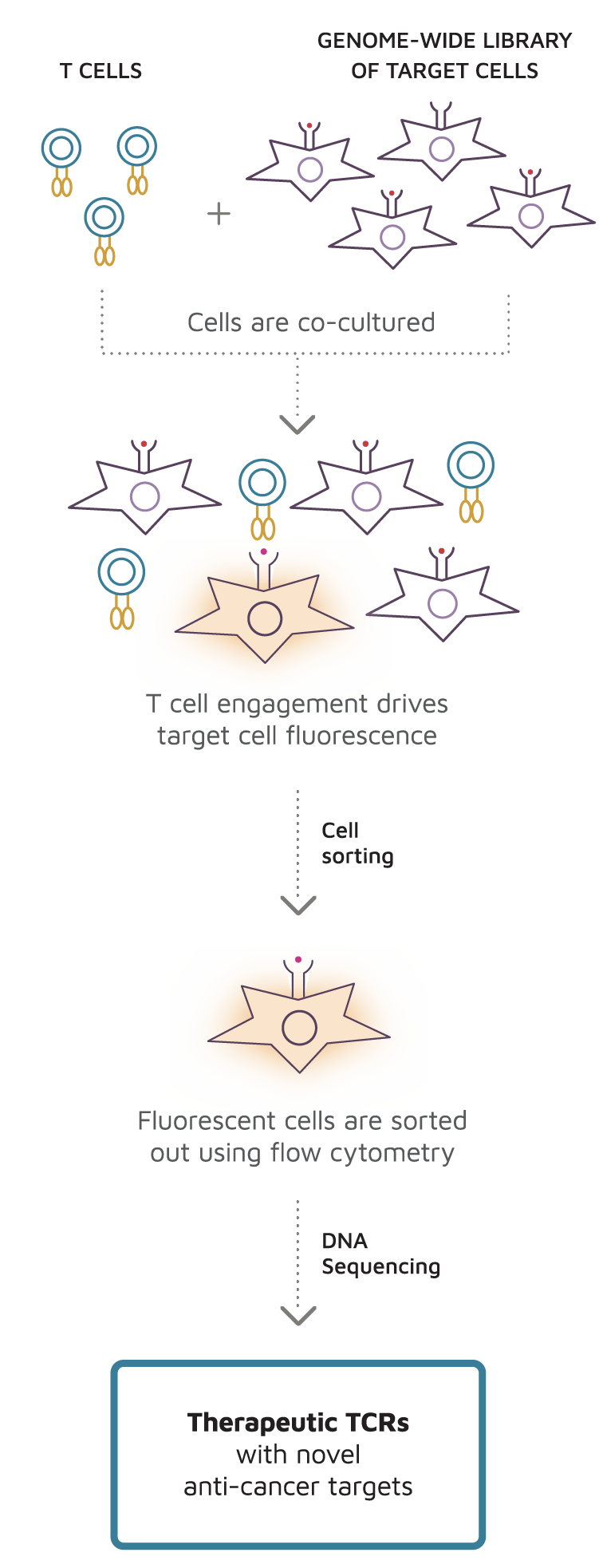
Overview of the TargetScan discovery process:
- T cells expressing a TCR of interest are co-cultured with a genome-wide library of target cells where every cell in the library expresses a different protein fragment. Each protein fragment is processed naturally by the proteasome or immunoproteasome and the resulting peptides are displayed on cell-surface major histocompatibility complex (MHC) proteins.
- If a T cell recognizes the peptide-MHC complex on a target cell, it attempts to kill the target cell, activating a proprietary fluorescent reporter in the target cell.
- By isolating fluorescent target cells and sequencing their expression cassettes, TargetScan reveals the natural target(s) of the T cell.
See Publications for the original article published in Cell in 2019.